We are interested in the functions of guanine nucleotide exchange factors (GEFs), a group of proteins known as activators of small G proteins. Our focus is on several GEFs for ARF, Rac and Rab GTPases. The goal is to find inhibitors which then allow us to characterize the cellular functions of the corresponding GEFs. A successful example of this approach is the cytohesin inhibitor SecinH3. Using this inhibitor we found a novel function of cytohesins as cytoplasmic activators of insulin signaling. Building on these findings a major current focus of the group is to elucidate the molecular mechanism by which cytohesins activate insulin signaling. The general approach in all the cell signaling projects is to integrate chemistry, chemical biology, biochemistry, structural biology and cell biology and to cover a wide spectrum of experimental systems from cell-free assays over cell culture models to in vivo applications. Although we select our targets with respect to their involvement in important physiological and pathophysiological processes, our primary goal is not the development of therapeutics but to use chemical biology to gain insight into fundamental cellular signaling processes.
An exciting new and rapidly emerging interdisciplinary field of research that merges the life- and engineering sciences in the discipline of Synthetic Biology. To this end, we have recently started to combine our expertise in aptamer research with DNA nanotechnology. Because of DNA's programmability and structural robustness, defined DNA structures like DNA rotaxanes and catenanes are an exciting new approach that promises to open an new field combining the areas of DNA nanotechnology and of interlocked molecular architectures. DNA rotaxanes and catenanes serve as model subtrates to construct and analyze DNA architectures containing moveable parts. We aim to construct DNA architectures that can exert defined functions. One important step to execute function is to be able to switch between different states of the DNA construct. We have developed diverse switches, based on orthogonal techniques. Structural rearrangements can be triggered either by light-irradiation, by toehold-extended nucleotides or by pseudo-complementary PNA (peptides nucleic acid).
DNA nanoarchitectures are analysed by HPLC, gel electrophoresis, and other biochemical techniques. Structures will be visualized by atomic force microscopy (AFM). We conduct our AFM studies in cooperation with Stefan Jester and Julián Valéro from the Kekulé Institute (University of Bonn). Because AFM is limited to DNA structures that adsorb on a surface, we now cooperate with Stephan Irsen (Electron Microscopy and Analytics, caesar) and Elmar Behrmann (Structural Dynamics of Proteins, caesar) on cryo-EM of DNA nanoarchitecturs to visualise larger DNA constructs.
The biochemical and AFM studies of this project are funded by the ERC Advanced Grant #267173.
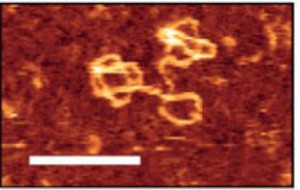
Figure: High-resolution image of NDA pseudorotaxanes. DNA pseudo-rotaxane constructs were assembled and HPLC-purified. DNA constructs were adsorbed on mica surfaces and visualized via atomic force microscopy. Areas whre DNA is ordered in layers give a brighter signal. Scale bar: 50 nm.
References
Ackermann, D. & Famulok, M. (2013) "Pseudo-complementary PNA actuators as reversible switches in dynamic DNA nanotechnology" Nucleic Acids Res. 41, 4729-4739
Li, T., Lohmann, F. & Famulok, M. (2014) "Interlocked DNA nanostructures controlled by a reversible logic circuit" Nat. Commun. 5, 4940
Lohmann, F., Weigandt, J., Valero, J. & Famulok, M. (2014) "Logic gating by macrocycle displacement using a double-stranded DNA [3]rotaxane shuttle" Angew. Chem. Int. Ed. Engl.53, 10372-10376